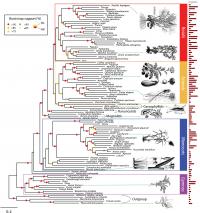
Scientists at the American Museum of Natural History, Cold Spring Harbor Laboratory, The New York Botanical Garden, and New York University have created the largest genome-based tree of life for seed plants to date. Their findings, published today in the journal PLoS Genetics, plot the evolutionary relationships of 150 different species of plants based on advanced genome-wide analysis of gene structure and function. This new approach, called "functional phylogenomics," allows scientists to reconstruct the pattern of events that led to the vast number of plant species and could help identify genes used to improve seed quality for agriculture.
more
Exploring science is typically characterized by a lot of puzzles, frustrations or even failures. This weblog is mainly intended to record my working, thinking and knowledge acquisitions. I expect that some reflection would refresh my mind from time to time, and motivate me to move further, and hopefully give me a better view about even changing the landscape of bioinformatics. You are welcome to leave some comments, good or bad, but hopefully something constructive. Enjoy your surfing!

Tuesday, December 27, 2011
Friday, December 9, 2011
KABOOM! A new suffix array based algorithm for clustering expression data
Abstract
Motivation: Second-generation sequencing technology has reinvigorated research using expression data, and clustering such data remains a significant challenge, with much larger datasets and with different error profiles. Algorithms that rely on all-versus-all comparison of sequences are not practical for large datasets.
Results: We introduce a new filter for string similarity which has the potential to eliminate the need for all-versus-all comparison in clustering of expression data and other similar tasks. Our filter is based on multiple long exact matches between the two strings, with the additional constraint that these matches must be sufficiently far apart. We give details of its efficient implementation using modified suffix arrays. We demonstrate its efficiency by presenting our new expression clustering tool, wcd-express, which uses this heuristic. We compare it to other current tools and show that it is very competitive both with respect to quality and run time.
Availability: Source code and binaries available under GPL athttp://code.google.com/p/wcdest. Runs on Linux and MacOS X.
Labels:
algorithm,
clustering,
software
Saturday, December 3, 2011
The Infobiotics Workbench: an integrated in silico modelling platform for Systems and Synthetic Biology
Abstract
Summary: The Infobiotics Workbench is an integrated software suite incorporating model specification, simulation, parameter optimization and model checking for Systems and Synthetic Biology. A modular model specification allows for straightforward creation of large-scale models containing many compartments and reactions. Models are simulated either using stochastic simulation or numerical integration, and visualized in time and space. Model parameters and structure can be optimized with evolutionary algorithms, and model properties calculated using probabilistic model checking.
Availability: Source code and binaries for Linux, Mac and Windows are available at http://www.infobiotics.org/infobiotics-workbench/; released under the GNU General Public License (GPL) version 3.
Labels:
software,
systems biology,
tools
Subscribe to:
Posts (Atom)