Exploring science is typically characterized by a lot of puzzles, frustrations or even failures. This weblog is mainly intended to record my working, thinking and knowledge acquisitions. I expect that some reflection would refresh my mind from time to time, and motivate me to move further, and hopefully give me a better view about even changing the landscape of bioinformatics. You are welcome to leave some comments, good or bad, but hopefully something constructive. Enjoy your surfing!

Showing posts with label SILAC. Show all posts
Showing posts with label SILAC. Show all posts
Sunday, February 27, 2011
Super-SILAC Technology for Quantitative Proteomics in Neoplasms
A group of investigators under Dr. Matthias Mann at Max Planck Institute of Biochemistry in Martinsried, Germany is working on an interesting quantitative proteomics technology that might offer a new way to analyze cell proteins in a range of disorders, such as cancer and autoimmune diseases. Called super-SILAC (stable-isotope labeling by amino acids in cell culture), the method generates thousands of isotopically labeled peptides in unique amounts to serve as "internal standards for mass spectrometry-based analysis."


Labels:
medical science,
proteomics,
quantitative proteomics,
SILAC
Thursday, February 3, 2011
Uniquant: an alternative to MaxQuant
UNiquant, a Program for Quantitative Proteomics Analysis Using Stable Isotope Labeling
Abstract

Stable isotope labeling (SIL) methods coupled with nanoscale liquid chromatography and high resolution tandem mass spectrometry are increasingly useful for elucidation of the proteome-wide differences between multiple biological samples. Development of more effective programs for the sensitive identification of peptide pairs and accurate measurement of the relative peptide/protein abundance are essential for quantitative proteomic analysis. We developed and evaluated the performance of a new program, termed UNiquant, for analyzing quantitative proteomics data using stable isotope labeling. UNiquant was compared with two other programs, MaxQuant and Mascot Distiller, using SILAC-labeled complex proteome mixtures having either known or unknown heavy/light ratios. For the SILAC-labeled Jeko-1 cell proteome digests with known heavy/light ratios (H/L = 1:1, 1:5, and 1:10), UNiquant quantified a similar number of peptide pairs as MaxQuant for the H/L = 1:1 and 1:5 mixtures. In addition, UNiquant quantified significantly more peptides than MaxQuant and Mascot Distiller in the H/L = 1:10 mixtures. UNiquant accurately measured relative peptide/protein abundance without the need for postmeasurement normalization of peptide ratios, which is required by the other programs.
Keywords (keywords):
Quantitative proteomics; Stable isotope labeling; LC-MS/MS; Software Development
Labels:
algorithm,
LC/MS,
quantitative proteomics,
SILAC,
software
Tuesday, November 16, 2010
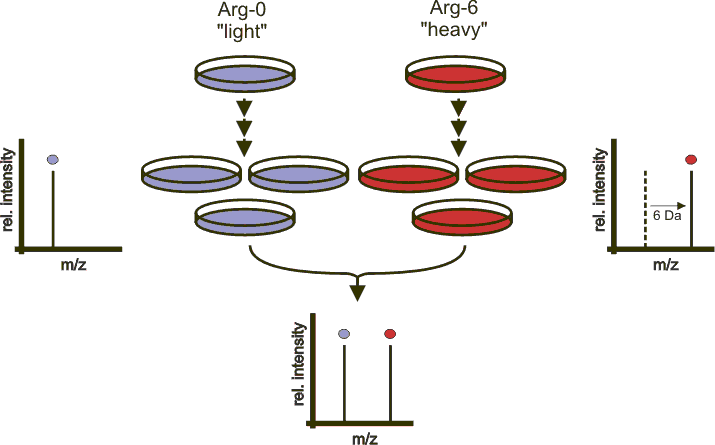
SILAC: Stable Isotope Labeling with Amino acid in Cell culture
SILAC is a simple and straight forward approach for in vivo incorporation of a label into proteins for MS-based quantitative proteomics. SILAC relies on metabolic incorporation of a given "light" or "heavy" form of the amino acid into proteins. The method relies on the incorporation of amino acids with substituted stable isotopic nuclei (e.g. deuterium, 13C, 15N). This in an experiment, two cell populations are grown in cultre media that are identical except that one of them contains a "light" and the other a "heavy" form of a particular amino acid (e.g. 12C and 13C labeled L-lysine, respectively).When the labeled analog of an amino acid is supplied to cells in culture instead of the natural amino acid, it is incorporated into all synthesized proteins. After a number of cell division, each instance of this particular amino acid will be replaced by its isotope labeled analog. Since there is hardly any chemical difference between the labeled amino acid and the natural amino acid isotopes, the cells behave exactly like the control cell population grown in the presence of normal amino acid. It is efficient and reproducible as the incorporation of the isotope is 100%.We anticipate of SIALC will lead to is use as a routine technology in all areas of cell biology.
The above is from http://silac.org/index_html
Labels:
mass spectrometry,
quantitative proteomics,
SILAC
Subscribe to:
Posts (Atom)