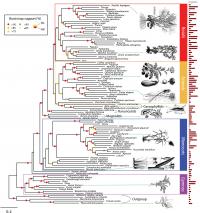
Scientists at the American Museum of Natural History, Cold Spring Harbor Laboratory, The New York Botanical Garden, and New York University have created the largest genome-based tree of life for seed plants to date. Their findings, published today in the journal PLoS Genetics, plot the evolutionary relationships of 150 different species of plants based on advanced genome-wide analysis of gene structure and function. This new approach, called "functional phylogenomics," allows scientists to reconstruct the pattern of events that led to the vast number of plant species and could help identify genes used to improve seed quality for agriculture.
more
Protein Life: R & D experience of a bioinformatician
Exploring science is typically characterized by a lot of puzzles, frustrations or even failures. This weblog is mainly intended to record my working, thinking and knowledge acquisitions. I expect that some reflection would refresh my mind from time to time, and motivate me to move further, and hopefully give me a better view about even changing the landscape of bioinformatics. You are welcome to leave some comments, good or bad, but hopefully something constructive. Enjoy your surfing!

Tuesday, December 27, 2011
Friday, December 9, 2011
KABOOM! A new suffix array based algorithm for clustering expression data
Abstract
Motivation: Second-generation sequencing technology has reinvigorated research using expression data, and clustering such data remains a significant challenge, with much larger datasets and with different error profiles. Algorithms that rely on all-versus-all comparison of sequences are not practical for large datasets.
Results: We introduce a new filter for string similarity which has the potential to eliminate the need for all-versus-all comparison in clustering of expression data and other similar tasks. Our filter is based on multiple long exact matches between the two strings, with the additional constraint that these matches must be sufficiently far apart. We give details of its efficient implementation using modified suffix arrays. We demonstrate its efficiency by presenting our new expression clustering tool, wcd-express, which uses this heuristic. We compare it to other current tools and show that it is very competitive both with respect to quality and run time.
Availability: Source code and binaries available under GPL athttp://code.google.com/p/wcdest. Runs on Linux and MacOS X.
Labels:
algorithm,
clustering,
software
Saturday, December 3, 2011
The Infobiotics Workbench: an integrated in silico modelling platform for Systems and Synthetic Biology
Abstract
Summary: The Infobiotics Workbench is an integrated software suite incorporating model specification, simulation, parameter optimization and model checking for Systems and Synthetic Biology. A modular model specification allows for straightforward creation of large-scale models containing many compartments and reactions. Models are simulated either using stochastic simulation or numerical integration, and visualized in time and space. Model parameters and structure can be optimized with evolutionary algorithms, and model properties calculated using probabilistic model checking.
Availability: Source code and binaries for Linux, Mac and Windows are available at http://www.infobiotics.org/infobiotics-workbench/; released under the GNU General Public License (GPL) version 3.
Labels:
software,
systems biology,
tools
Saturday, November 26, 2011
Nano-ultra high performance liquid chromatography
Thermo Fisher Scientific Inc says the Center for Experimental Bioinformatics (CEBI) at the University of Southern Denmark is using Thermo Scientific nano-ultra high performance liquid chromatography (UHPLC) technology to accelerate productivity and improve efficiencies during its proteomics research.
Implementation of the Thermo Scientific EASY-nLC 1000 has enabled 34 percent more peptide and 22 percent more protein identifications within the CEBI laboratory, significantly increasing productivity. Additionally, the instrument has improved separations and facilitated the analysis of more complex samples within the laboratory.
Based in Odense, the University of Southern Denmark’s Department of Biochemistry and Molecular Biology is world-renowned in the field of proteomics, with research focusing on gene structure and organization, regulation and RNA translation. CEBI is a specialist proteomics research laboratory, aimed at applying modern methods of mass spectrometry and proteomics to functional analysis of genes.
Prior to implementing nano-UHPLC technology, CEBI used conventional HPLC. However they needed a more complete solution, such as EASY-nLC 1000, to accelerate productivity and improve efficiencies. The EASY-nLC 1000 is used in conjunction with Thermo Scientific ion trap and Orbitrap mass spectrometers for a wide range of proteomics research, including the study of complex mouse dendritic cells.
Using the EASY-nLC 1000 has allowed CEBI to take advantage of the ability to facilitate dedicated separation of biomolecules at ultra-high pressures. The higher operating pressures provided by the EASY-nLC 1000 have increased the number of peptide and protein identifications performed within the laboratory by 34 percent and 22 percent respectively. The instrument also allows selection of the most appropriate column dimensions and solid phase materials in order to identify the most suitable configuration to match analytical requirements. Using thinner, longer columns and smaller beads has improved separation and ionization, enhancing sensitivity and allowing the CEBI team to scale down experiments and reduce costs.
“The EASY-nLC 1000 is a highly robust instrument offering nano-UHPLC performance dedicated to proteomics scientists,” said Lasse Falkenby, research assistant at CEBI. “We decided to implement the new system in our laboratory as we felt that it would revolutionise the way we perform LC-MS analyses. The instrument has allowed our laboratory to increase productivity considerably in a way that would never have been possible before. Implementation was a very smooth process requiring minimal configuration effort and the Thermo Fisher Scientific team of experts provided valuable support both throughout and after this process.”
For more information, visit www.thermoscientific.com/easylc
Implementation of the Thermo Scientific EASY-nLC 1000 has enabled 34 percent more peptide and 22 percent more protein identifications within the CEBI laboratory, significantly increasing productivity. Additionally, the instrument has improved separations and facilitated the analysis of more complex samples within the laboratory.
Based in Odense, the University of Southern Denmark’s Department of Biochemistry and Molecular Biology is world-renowned in the field of proteomics, with research focusing on gene structure and organization, regulation and RNA translation. CEBI is a specialist proteomics research laboratory, aimed at applying modern methods of mass spectrometry and proteomics to functional analysis of genes.
Prior to implementing nano-UHPLC technology, CEBI used conventional HPLC. However they needed a more complete solution, such as EASY-nLC 1000, to accelerate productivity and improve efficiencies. The EASY-nLC 1000 is used in conjunction with Thermo Scientific ion trap and Orbitrap mass spectrometers for a wide range of proteomics research, including the study of complex mouse dendritic cells.
Using the EASY-nLC 1000 has allowed CEBI to take advantage of the ability to facilitate dedicated separation of biomolecules at ultra-high pressures. The higher operating pressures provided by the EASY-nLC 1000 have increased the number of peptide and protein identifications performed within the laboratory by 34 percent and 22 percent respectively. The instrument also allows selection of the most appropriate column dimensions and solid phase materials in order to identify the most suitable configuration to match analytical requirements. Using thinner, longer columns and smaller beads has improved separation and ionization, enhancing sensitivity and allowing the CEBI team to scale down experiments and reduce costs.
“The EASY-nLC 1000 is a highly robust instrument offering nano-UHPLC performance dedicated to proteomics scientists,” said Lasse Falkenby, research assistant at CEBI. “We decided to implement the new system in our laboratory as we felt that it would revolutionise the way we perform LC-MS analyses. The instrument has allowed our laboratory to increase productivity considerably in a way that would never have been possible before. Implementation was a very smooth process requiring minimal configuration effort and the Thermo Fisher Scientific team of experts provided valuable support both throughout and after this process.”
For more information, visit www.thermoscientific.com/easylc
Thursday, November 17, 2011
Pico Computing Demonstrates Bioinformatics Acceleration at SC 2011
Pico's SC5 FPGA cluster reduces short read sequencing from 6 1/2 hours to just one minute
Seattle, WA (PRWEB) November 09, 2011
Pico Computing will be demonstrating an FPGA implementation of BFAST resulting in a 350X acceleration over a software-only implementation running on two quad core Intel Xeon processors. The 350X acceleration was achieved using 8 Pico M-503 FPGA modules in their SC5 SuperCluster chassis. The BFAST algorithm is primarily used in short read genome mapping.
Read more: http://www.sfgate.com/cgi-bin/article.cgi?f=/g/a/2011/11/09/prweb8945226.DTL#ixzz1dyI5LQs
Labels:
next generation sequencing,
supercomputing
Monday, November 7, 2011
Gene Ontology-driven inference of protein-protein interactions using inducers
Motivation: Protein-protein interactions (PPI) are pivotal for many biological processes and similarity in Gene Ontology (GO) annotation has been found to be one of the strongest indicators for PPI. Most GO-driven algorithms for PPI inference combine machine learning and semantic similarity techniques. We introduce the concept of inducers as a method to integrate both approaches more effectively, leading to superior prediction accuracies.
Results: An inducer (ULCA) in combination with a Random Forest classifier compares favorably to several sequenced-based methods, semantic similarity measures and multi-kernel approaches. On a newly created set of high-quality interaction data, the proposed method achieves high cross-species prediction accuracies (AUC ≤ 0.88), rendering it a valuable companion to sequence-based methods.
Availability: Software and datasets are available athttp://bioinformatics.org.au/go2ppi/
Labels:
algorithms,
gene annotation,
genomics,
PPI,
software
Saturday, October 29, 2011
Enhanced peptide quantification using spectral count clustering and cluster abundance
Quantification of protein expression by means of mass spectrometry (MS) has been introduced in various proteomics studies. In particular, two label-free quantification methods, such as spectral counting and spectra feature analysis have been extensively investigated in a wide variety of proteomic studies.
The cornerstone of both methods is peptide identification based on a proteomic database search and subsequent estimation of peptide retention time. However, they often suffer from restrictive database search and inaccurate estimation of the liquid chromatography (LC) retention time.
Furthermore, conventional peptide identification methods based on the spectral library search algorithms such as SEQUEST or SpectraST have been found to provide neither the best match nor high-scored matches. Lastly, these methods are limited in the sense that target peptides cannot be identified unless they have been previously generated and stored into the database or spectral libraries.To overcome these limitations, we propose a novel method, namely Quantification method based on Finding the Identical Spectral set for a Homogenous peptide (Q-FISH) to estimate the peptide's abundance from its tandem mass spectrometry (MS/MS) spectra through the direct comparison of experimental spectra.
Intuitively, our Q-FISH method compares all possible pairs of experimental spectra in order to identify both known and novel proteins, significantly enhancing identification accuracy by grouping replicated spectra from the same peptide targets.
Results: We applied Q-FISH to Nano-LC-MS/MS data obtained from human hepatocellular carcinoma (HCC) and normal liver tissue samples to identify differentially expressed peptides between the normal and disease samples. For a total of 44,318 spectra obtained through MS/MS analysis, Q-FISH yielded 14,747 clusters.
Among these, 5,777 clusters were identified only in the HCC sample, 6,648 clusters only in the normal tissue sample, and 2,323 clusters both in the HCC and normal tissue samples. While it will be interesting to investigate peptide clusters only found from one sample, further examined spectral clusters identified both in the HCC and normal samples since our goal is to identify and assess differentially expressed peptides quantitatively.
The next step was to perform a beta-binomial test to isolate differentially expressed peptides between the HCC and normal tissue samples. This test resulted in 84 peptides with significantly differential spectral counts between the HCC and normal tissue samples.
We independently identified 50 and 95 peptides by SEQUEST, of which 24 and 56 peptides, respectively, were found to be known biomarkers for the human liver cancer. Comparing Q-FISH and SEQUEST results, we found 22 of the differentially expressed 84 peptides by Q-FISH were also identified by SEQUEST.
Remarkably, of these 22 peptides discovered both by Q-FISH and SEQUEST, 13 peptides are known for human liver cancer and the remaining 9 peptides are known to be associated with other cancers.
Conclusions: We proposed a novel statistical method, Q-FISH, for accurately identifying protein species and simultaneously quantifying the expression levels of identified peptides from mass spectrometry data. Q-FISH analysis on human HCC and liver tissue samples identified many protein biomarkers that are highly relevant to HCC.
Q-FISH can be a useful tool both for peptide identification and quantification on mass spectrometry data analysis. It may also prove to be more effective in discovering novel protein biomarkers than SEQUEST and other standard methods.
Author: Seungmook LeeMin-Seok KwonHyoung-Joo LeeYoung-Ki PaikHaixu TangJae LeeTaesung Park
Credits/Source: BMC Bioinformatics 2011, 12:423
Subscribe to:
Posts (Atom)